Raw sequences produced by next generation sequencing (NGS) machines may contain adapter, linker, barcode and fingerprint sequences. TagDust2 is a program to extract and correctly label the sequences to be mapped in downstream pipelines.
TagDust allows users to specify the expected architecture of a read and converts it into a hidden Markov model. The latter can assign sequences to a particular barcode (or index) even in the presence of sequencing errors. Sequences not matching the architecture (primer dimers, contaminants etc.) are automatically discarded:
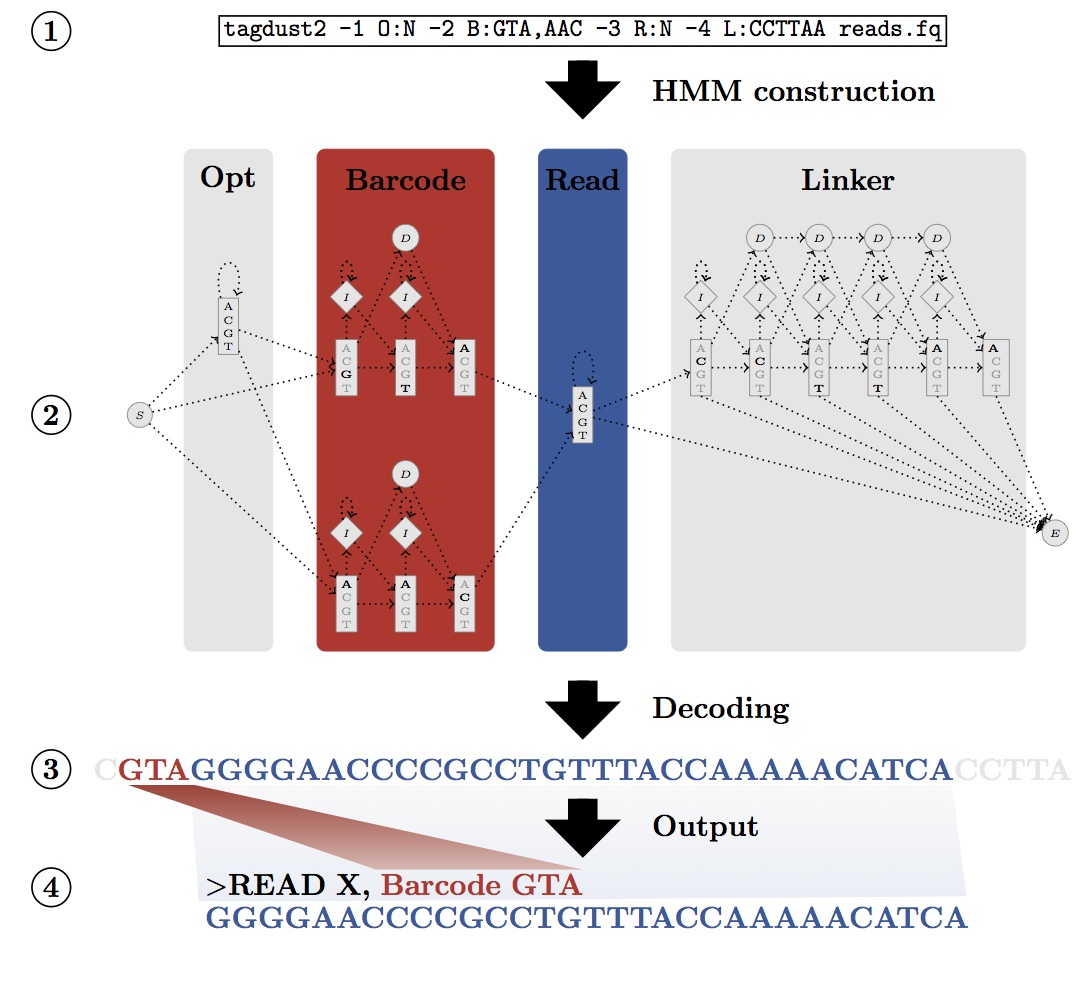
Version 2.2 is about 2 major updates
1) Integration with Illumina Casava output.
2) A test suite was added to ensure all paper figures can be easily reproduced.
Have a look at the expanded user manual.
Version 2.12 includes minor bug fixes.
Version 2.11 includes three major updates:
1) Support for both single and paired end data.
2) Automatic selection of read architectures. In a lab running several NGS protocols users can now create a template file listing all possible read architectures / barcode combinations used. TagDust will automatically select the correct architecture.
3) Automatic threshold selection. In pervious versions the -q option was used to select the extraction threshold. The new version runs a small simulation and automatically selects a threshold giving best sensitivity + specificity. This greatly improves the results when the read architecture is simple (e.g. a three letter barcode followed by the read).
tar -zxcf tagdust-X.XX.tar.gz
cd tagdust
./configure
make
make check
make install
